why
We all know that roots are the organ system that gives plants some structure and allows them to access nutrients and water, affecting the hydrological cycle. Since the distribution of these resources is non-uniform in the soil, the position, shape, and density of plants’ roots influences their uptake efficiency. Less popularly, roots also produce metabolites that help make soil the planet’s store of carbon by excellence.
Isn’t it fascinating that despite lacking a centralized information processing center (like our nervous system), plants joggle responses to several complex environmental inputs? How could we use this knowledge to help them make a more efficient use of their resources with a sustainable agriculture vision in mind?
When it comes to optimizing, let alone designing, these roots, we’ve been limited by our ability to design and build predictable genetic systems in plants, especially when no native promoters are available. Unlike other organisms, plants have proven to need longer timelines and it’s been hard to tune circuit activity across heterogenous plant cells.
This study thus develops quantitative transient expression assays as a platform to test and tune genetic circuits in plants.
what
As a refresher, transcription factors (TFs) are molecules that help in the process of gene expression in humans and other eukaryotes like plants. General TFs aid in the overall binding of RNA polymerase to DNA while specific ones aid the former ones in the binding to specific promoters.
When thinking of genetic logic you can imagine a specific combination of transcriptional regulators (a.k.a, TF) needed to unlock, or lock, the expression of genes. Synthetic biology comes in when we want to intelligently design and express the logic we want in living things or its parts.
This paper specifically, builds synthetic transcriptional regulators that can be compiled to create genetic circuits that perform Boolean logic:
transcriptional activators: bacterial DNA-binding proteins + 6x VP16 activation domains (operators) + SV40 NLSs.
9/10 turned on their target promoters
7 were specific to their target
transcriptional repressors: @ the end of CaMV 35S promoter (strong constituitive promoter in plants)
4/10 had a >2x change in gene expression
notes: mCherry was used to normalize the fluorescence measurements (as a point of reference). Promoters were also engineered to tune expression levels by changing the # of transcription factors binding sites (operators) in the AmtR promoter and ≥3 was the ideal number. Location of the operator also affected repression and dynamic range.
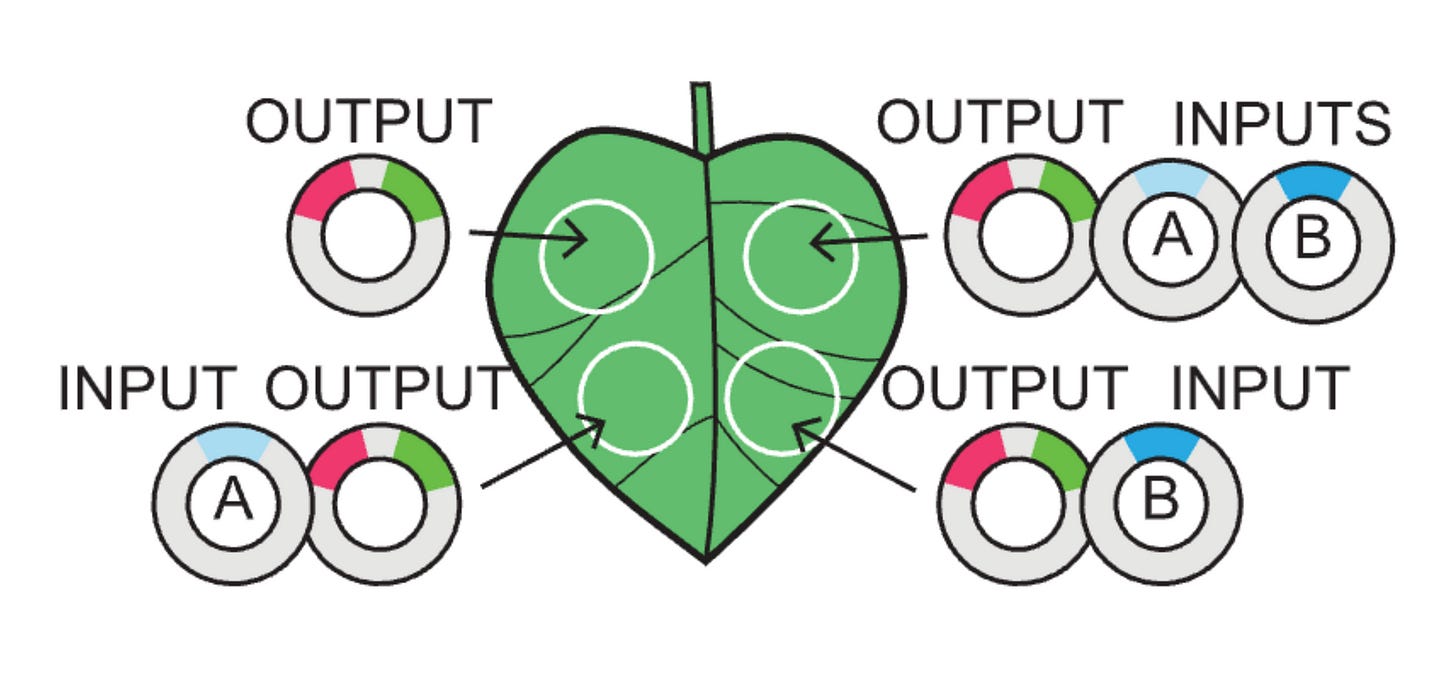
The actual logic gates and genetic circuits were built in the following way:
input: plasmid with a transcriptional regulator like an activator, a repressor, or both.
output: plasmid with recognition sites for the input(s) + specific promoter + gene of interest
Keep reading with a 7-day free trial
Subscribe to Biopunk to keep reading this post and get 7 days of free access to the full post archives.